Plasmid genomics, antibiotic resistance, and environmental pathogens
In the Herrick laboratory, we use genomic and microbiological techniques to study populations of native and introduced -- including pathogenic -- bacteria in natural streams and soils. Our main interest is in the lateral transfer of genes to and among bacteria in streams and soils. Lateral gene transfer - as opposed to the 'vertical' transfer of genes via simple cell division - allows genes to move between mature cells and thus to potentially spread very quickly through a population and even from species to species. Lateral gene transfer has had a profound effect on genome evolution, on pollutant biodegradation, and particularly on the development of antibiotic resistance in bacteria.
Our current focus is on the transmission of plasmid-borne resistance genes in stream waters and sediments. Antibiotic overuse and misuse may lead to selection for resistance genes, many of which are found on mobile genetic elements such as plasmids and transposons, and these can potentially be transferred from human or animal strains to bacteria in the environment. Resistant environmental bacterial populations may then act as environmental reservoirs and evolutionary "incubators" of resistance genes, thus providing new variants and combinations of resistance phenotypes for subsequent transfer to human and animal pathogenic bacteria. We are currently using genetic 'capture' techniques and comparative genomics methods to study actively transferring resistance plasmids which, though acquired from native stream bacteria, may have their origins in introduced, antibiotic-selected fecal bacteria.
We are also using whole genome sequencing -- both short-read and long-read -- and comparative genomics to study pathogenic bacteria, especially Salmonella, in streams. Students have isolated numerous Salmonella strains from stream sediments throughout the Shenandoah Valley, some of which are very close relatives to serotypes implicated in recent outbreaks. Our particular interest is in their mobile genetic elements -- especially plasmids and transposons -- and the potential these have for transferring virulence and resistance genes to and from native stream populations.
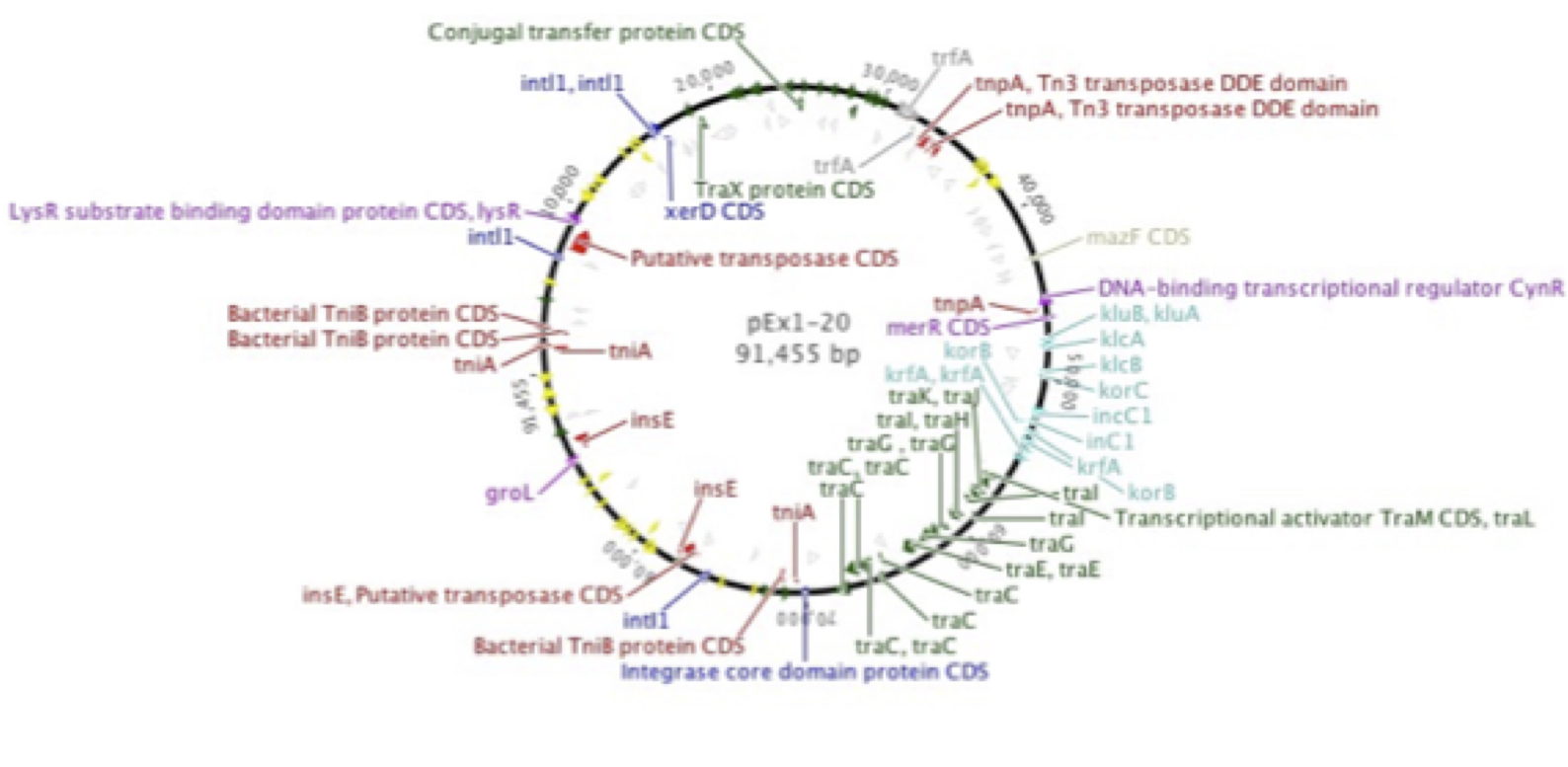
Plasmid p1-20, isolated by graduate student Erika Gehr from Shull's Run, a stream in the Shenandoah Valley, was found to encode resistance to a number of antibiotics, many of which are late-generation, clinical use-only drugs. This supported earlier work in our laboratory (Herrick et al. 2014. Coselection for resistance to multiple late-generation human therapeutic antibiotics encoded on tetracycline resistance plasmids captured from uncultivated stream and soil bacteria. Journal of Applied Microbiology, 117, 380-389) and was confirmed by graduate student Kevin Libuit by DNA sequencing. Numerous genes encoding resistance to beta-lactamases, aminoglycosides, tetracyclines, etc., in addition to mobile elements such as multiple transposons and integrons were found.

The MinION nanopore DNA sequencer
The Herrick laboratory was one of the first in the world, as part of the Oxford Nanopore MAP program, to use the MinION DNA sequencer. The sequencer is smaller than a harmonica, extremely fast, and easy to use. We have used it, both alone and in conjunction with the CGEMS Ion Torrent PGM DNA sequencer, to sequence multiresistance plasmids captured, without cultivation, from stream sediment.
